by Katie Sinclair
Katrina Lohan and Kristy Hill have travelled thousands of miles down the Atlantic Coast, from the Chesapeake to the Caribbean. Their goal? Track the range and distribution of parasites in bivalve mollusks that could cause disease. Based on diversity patterns, Hill and Lohan suspect that there are many more protist species in the tropics than have previously been discovered. These parasites could be very similar to the parasites that have caused mass die-offs in Chesapeake oyster beds with diseases like Dermo and MSX.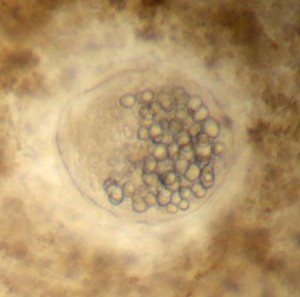
Close-up of a trematode oyster parasite. These parasites form cysts, and could be similar to the parasites that caused mass die-offs in the Chesapeake.
Lohan, a postdoc who works jointly with Greg Ruiz at SERC and with Robert Fleischer at the Smithsonian Conservation Biology Institute at the National Zoological Park, and Hill of SERC are using DNA sequences to identify bivalve parasites. Knowing exactly which species is infecting each oyster is important, according to Lohan, because it can tell researchers a lot about parasite ecology and offer clues about disease transmission.
DNA sequencing is one of the few tools that can tell biologists what protists are out there. Besides taking oyster tissue samples, Hill and Lohan also sequence DNA from water samples. Identifying a single genus from thousands of microscopic organisms is challenging, but comparing the parasites found in oyster tissues to those found in the water can shed more light on parasites’ roles in the ecosystem. Protist genomes have not been studied extensively, so Lohan and Hill limit their search to a few key genes. “Many have weird genomes that haven’t been sequenced yet,” Lohan said.
Conserving an Endangered Orchid
Melissa McCormick and Dennis Whigham certainly understand the allure of new species with weird genomes. Although both are plant ecologists, they’ve spent years researching fungi. Over 90 percent of all terrestrial plants rely on symbiotic relationships with fungi—termed mycorrhizal fungi—to extract the nutrients they need to survive. SERC holds the largest collection of orchid mycorrhizal fungi in the world. Understanding how those fungi are distributed could help efforts to conserve and protect endangered orchids, including the small-whorled pogonia, one of the most endangered orchid species in North America.Because orchid mycorrhizal fungi often lack structures that allow them to be identified by morphology, DNA sequencing is one of the few ways to identify a fungus species. “Only about 17 species of this type of fungi have been described, but DNA suggests that there’s over 100 different types,” said McCormick, who runs SERC’s Molecular Ecology Lab.
McCormick’s work on fungi has also led her to investigate the little-understood realm of bacteria. It’s possible that these fungi need particular bacteria within their cells to be able to help orchids germinate. By extracting DNA from bacteria that live in mycorrhizal fungi, the lab may be on the cusp of identifying previously undescribed species.
“Just seeing what we suspect to be endobacteria in orchid mycorrhizal fungi was really cool and unexpected,” said Wes Hauser, a Wabash College undergraduate interning with Dennis Whigham in the Plant Ecology Lab this summer. “Nobody has had the sort of collections that Melissa and Dennis have at SERC, so we’re some of the first ones to look at endobacteria in orchid mycorrhizal fungi.” Unlocking the secrets of the endobacteria could take them one step closer to bringing the small-whorled pogonia back.
Understanding Invaders
Besides orchids, the Plant Ecology Lab and Molecular Ecology Lab also use DNA to track the spread of the invasive reed Phragmites australis. Using “genetic fingerprints” can help determine how fast a particular genotype of Phragmites is spreading. Comparing chloroplast DNA to nuclear DNA can show whether invasive Phragmites is spreading through pollen or seeds.
“It can help us identify a ‘tipping point’ in a watershed—how close two different genotypes have to be in order to produce seeds, how much Phragmites you have to remove in order to protect the region you’re managing,” McCormick said.
To uncover how invasive blue catfish impact food webs, the Fish and Invertebrate Lab uses DNA to identify the fish’s stomach contents. Distinguishing different fish species from blobs of half-digested goo can be nearly impossible, so the arrival of DNA identification techniques is a much less messy method to figure out who’s eating whom.
DNA is used to identify intact organisms as well. “Barcoding,” a technique which looks at sequences only several hundred base pairs long, is a fast and effective way to identify species. Eventually, the Fish and Invertebrate Lab hopes to create a comprehensive database of all the fish and macroinvertebrate species found in the Chesapeake Bay and its tributaries.
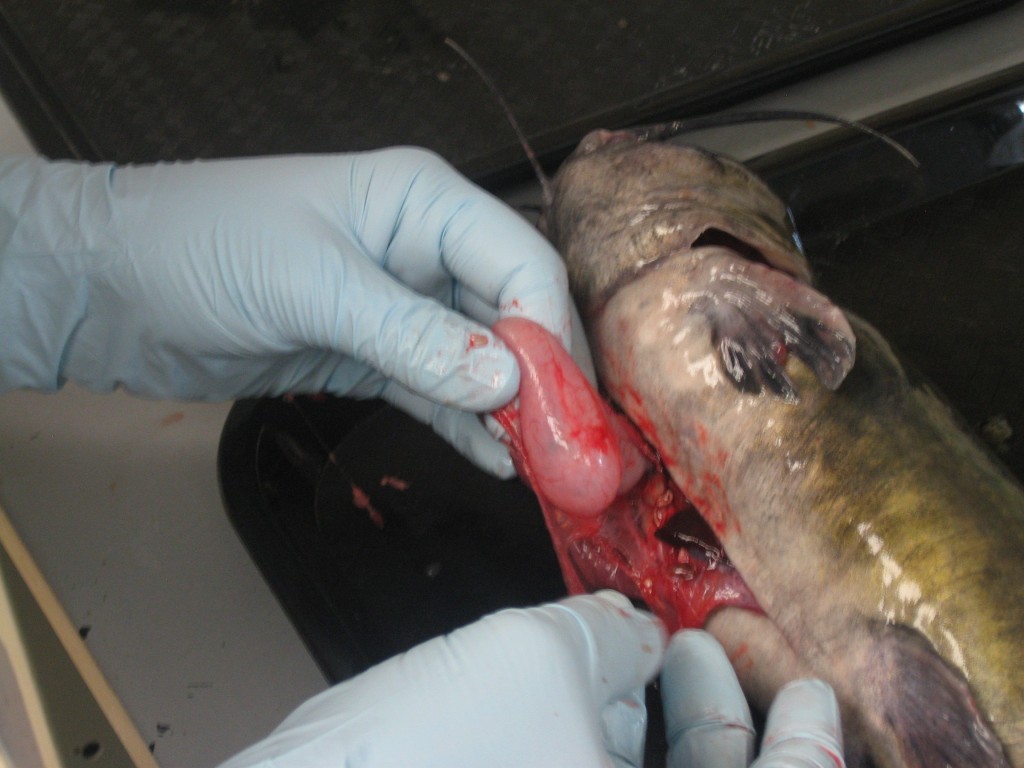
Removing a catfish stomach is messy work, but it’s one of the best ways to figure out who’s eating whom. (SERC)
But despite becoming more and more common, DNA is still a topic that many people are unfamiliar with outside the fields of medicine and forensic science.
“I think a lot of the public feels—like me, before working with genetics—that it’s something hard to grasp. But the information we can get out of it is so important,” Hill said.
Ultimately, DNA can be a valuable tool when conserving and managing rare and endangered species. But the main allure of DNA is the sheer amount of information it can reveal. One single sequence can provide an extraordinary amount of information. DNA can help discover new species, and—in Hill’s case—discover species in new places. “I feel like Lewis and Clark almost, going out to these places and finding things that no one’s ever found before.”




Hooray for DNA analysis. Looks like an interesting job, and thanks for the infomation:)