by Kristen Minogue

Blue catfish SERC biologists dubbed “Megalodon,” which they tracked moving almost 60 miles along the Patuxent River. (Brooke Weigel/SERC)
White perch, menhaden and darters: These are just a few favorite foods of Maryland’s invasive blue catfish, according to a new study from the Smithsonian Environmental Research Center (SERC). They’re also known to gorge themselves on larvae of channel catfish—and, occasionally, juveniles of their own kind.
The study, published in the journal Environmental Biology of Fishes, used DNA barcoding to get to the gut of what blue catfish prey on. Blue catfish arrived in Chesapeake Bay in the 1960s, brought by Virginia managers to establish a fishery. They quickly developed a reputation as voracious predators, threatening to devour many popular fisheries and edge out the Chesapeake’s native white catfish. However, to discover how much they could disrupt the ecosystem, marine biologists need to know exactly what they eat. The only way to do that is to look into their stomachs, where the majority of their prey has been reduced to almost-unrecognizable slop.
Rob Aguilar would know: A biologist with SERC’s Fish and Invertebrate Lab, he’s spent the last few years dissecting blue catfish stomachs and analyzing their insides.
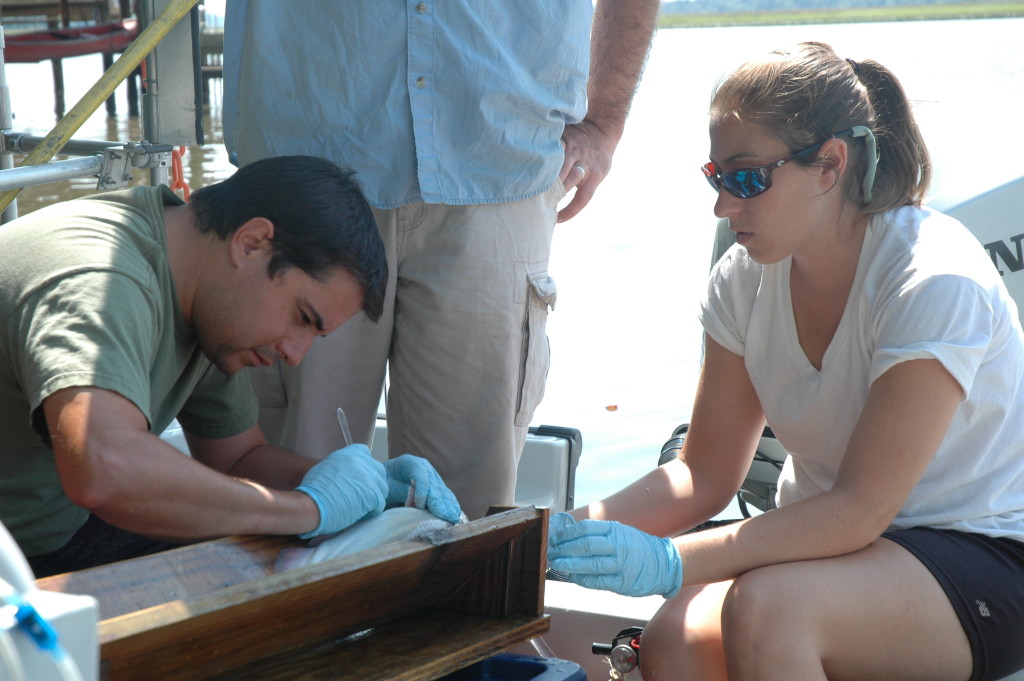
Rob Aguilar (left) and Paige Roberts perform surgery on a blue catfish. In addition to analyzing their stomachs, the Fish & Invertebrate Lab is also tagging blue catfish to track their movements. (Brooke Weigel/SERC)
“Most of the time the fish was fairly well digested, so it was hard to tell exactly what it was,” Aguilar said. “You would see maybe some tissue on a spine, some loose tissue, chunks of fish. You knew it was a fish, but that was the best you could do.”
Podcast: Rob Aguilar talks about using DNA to identify the contents of fish guts
DNA barcoding offered a way to identify what species the tissues came from. Barcoding involves extracting a tiny DNA fragment from the digested tissue and looking it up in a DNA library. The two DNA libraries the team used, called GenBank and the Barcode of Life Database (BOLD), contain DNA sequences for hundreds of thousands of species, including hundreds from the Chesapeake. Many of the libraries’ Chesapeake samples were added by Aguilar and the lab as part of the Chesapeake Bay Barcode Initiative, a project to gather DNA from all the Bay’s fish and major invertebrates. As of October 2016, they’ve collected DNA from two-thirds of its fish species—almost all the ones that don’t fall into the “extremely rare” category—and over 200 of the near-1,000 macroinvertebrates.
Learn more: DNA Barcodes Identify Chesapeake Species
All totaled, the lab dissected over 600 catfish stomachs and extracted DNA from more than 100 of them—mostly invasive blues, but a couple dozen native whites and channel catfish, a non-native but long-established catfish in Maryland.
Visually, the lab could identify to species about 9 percent of the fish items inside stomachs that also went through DNA analysis (not counting fish eggs). DNA barcoding nailed down 90 percent.
The wild success wasn’t a surprise to Matt Ogburn, a principal investigator in the Fish and Invertebrate Lab. “We figured it would work a lot better,” he said. “We know that the traditional methods of using morphology to identify gut contents can be really difficult.”
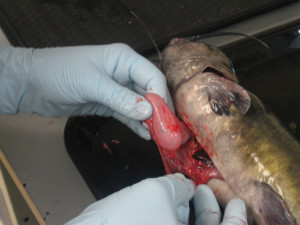
Blue catfish stomach (Katie Sinclair/SERC)
The surprises came in some of the species they found. The most popular food by sheer numbers was newly hatched larvae of channel catfish. Aguilar remembered dissecting stomachs crammed with hundreds of them, leading him to suspect blue catfish were raiding channel catfish nests. Other common prey included white perch, tessellated darters and menhaden, tiny fish that aren’t popular as seafood but are immensely valuable for fish oils, cosmetics and crab bait.
But the lab also uncovered evidence of scavenging (eating dead animals), and cannibalism.

Fish spine from dissected stomach (Miranda Marvel/SERC)
Some fish scales in the catfish stomachs were so large, their owners were likely too big for blue catfish to eat whole, implying the animals were eating something already dead. Then there was the random tissue sample that came back as a double-crested cormorant. Ogburn thinks the bird almost certainly died first before landing in a spot where a blue catfish could grab a few quick bites.
Eight blue catfish stomachs also contained DNA from their own species mixed with other digested prey, a red flag that those catfish were eating their own kind, most likely smaller juveniles.

White perch remains taken from a catfish stomach. (SERC)
This study looked only at fish prey for blue catfish in Maryland. It’s a critical snapshot, the biologists say, because they’ve only been in Maryland two decades. In Virginia, where blue catfish have been since the 1960s, they’ve had time to grow larger and more numerous—and bigger blue catfish tend to eat more fish and little else.
“We could be at the forefront of that,” Aguilar said. “We don’t exactly know what’s going to happen, so it would be good to get some of this data as the populations of blue catfish increase.”
“They’re definitely changing things,” Ogburn said. “They get much bigger than our native catfish do, and they’re much more abundant. And we know some of what they‘re doing is eating other species that we care about.” On the other hand, he added, “They’re not going to wipe out all the blue crabs or all the striped bass or some of the things people have worried about.”
In the meantime, Aguilar, Ogburn and the rest of the lab will continue adding DNA from more Chesapeake species to the barcode library, to unravel more secrets hiding in the stomachs of the Bay’s predators.
Learn more:
Podcast: All About that Base…Pairs. Using DNA Barcoding to Identify Fish Gut Contents
Decoding Nature: How DNA Can Save Species
DNA Barcodes Identify Chesapeake Species
Chesapeake Bay Barcode Initiative Homepage

